Hacker Perspective: Folding@home Tips and Advocacy
In 1597, Francis Bacon wrote 'knowledge is power'. The Folding@home project is proof that 'power (both electrical + computer) produces knowledge'. This is even more true with the rise of Artificial Intelligence projects like Google's AlphaFold.

Misfolded proteins have been implicated in numerous diseases. Folding@home is biological research based upon the science of Molecular Dynamics where Molecular Chemistry and mathematics are combined in computer-based models to predict how protein molecules might fold, or misfold, in space over time. This information is then used to guide scientific and medical research.
When I first heard about this, I recalled the science-fiction magnum opus by Isaac Asimov colloquially known as The Foundation Trilogy which introduced the fictional science of psychohistory (where statistics, history and sociology were combined in computer-based models) to guide humanity's future in order to reduce a potential 30,000 year galactic dark age to only 1,000 years.
During my formative years, I became infected with an Asimov inspired optimism about humanity's future and have since felt the need to promote Asimov's vision. While Folding@home will not cure my "infection of optimism", I am convinced Dr. Isaac Asimov (PhD in Biochemistry from Columbia in 1948, then was employed as a Professor of Biochemistry at the Boston University School of Medicine for 10-years until his personal publishing workload became too large) would have been fascinated by something like this.
I was considering a financial charitable donation to Folding@home when it occurred to me that my money would be better spent by making a knowledgeable charitable donation to all of humanity by:
- Increasing my Folding@home computations (which will advance medical discoveries to increase both the length and quality of human life). I was already folding on a half-dozen computers so all I needed to do was purchase video graphics cards which would increase my computational throughput by a thousand-fold (three orders of magnitude).
- Showing you how to get the client working on Linux which produces faster results than Windows
- Convincing others (like you) to do run Folding@home on any platform. My solitary folding efforts will have little effect on humanity's future but together we can make a real difference.
Protein Folding Overview
Science Problem
Real-world observation
From a kitchen point of view, chicken eggs are a mix of water, fat (yolk), and protein (albumen). Cooking an egg causes the semi-clear protein to unfold into long strings which now can intertwine into a tangled network which will stiffen then scatter light (now appears white). No chemical change has occurred, but taste, volume and color have been altered.
Click here to read a short "protein article" by Isaac Asimov published in 1993 shortly after his death.
Two (of many) TED-Talk Videos:
Computer Solutions
Single CPU Systems
Using the most powerful single core processor (Pentium-4), simulating the folding possibilities of one large protein molecule for one millisecond of chemical time might require one million days (2737 years) of computational time. However, if the problem is sliced up then assigned to 100,000 personal computers over the internet, the computational time drops to ten days. Convincing friends, relatives, and employers to do the same could reduce the computational time requirement to one day or less.| chemical time in nature |
required simulation time | ||
|---|---|---|---|
| one computer |
100,000 computers |
1 million computers |
|
| 1 Second | 1 billion days (2.7 million years) |
27 years | 2.7 years |
| 1 mS | 1 million days (2,737 years) |
10 days | 1 day |
Additional information for science + technology nerds
Not a nerd? Click skip this section
- Special-purpose research computers like IBM's Blue Gene employ 10 to 20 thousand processors
(CPUs) joined by many kilometers of optical fiber to solve problems. IBM's Roadrunner is a similar technology employing both "CPUs" and "special non-graphic GPUs that IBM refers to as cell processors"
- The basic terms:
- Early CPUs were built around integer processing with floating point operations being simulated in software.
- Later CPUs supported both integer and floating point operations in hardware
- The combined throughput (fetch floating point data from memory, manipulate it, then write it back) is known as a FLOP (FLoating point OPeration) with the total spec being published as FLOPS (FLoating point OPerations per Second)
- As of June 2023, the Folding@home project consists of ~ 40,000 active platforms (some hosting 14,000 GPUs) which yield 26,135 TeraFLOPS (26 PetaFLOPS).
- Equivalents:
- A Pentium-4 was rated at 12 GigaFLOPS
(26 x 10^15) / (12 x 10^9) = 2,167,000 Pentium-4 processors - A Core i7 was rated at 100 GigaFLOPS which is 8-times higher than the Pentium-4).
(26 x 10^15) / (100 x 10^9) = 260,000 Core i7 processors
- A Pentium-4 was rated at 12 GigaFLOPS
- This means that the original million-day protein simulation problem could theoretically be completed in (1,000,000 / 2,167,000) 0.46 days (or 11 hours). But since there are many more protein molecules than DNA molecules, humanity could be at this for decades. Adding your computers to Folding@home will permanently advance humanity's progress in protein research and medicine.
- The basic terms:
- When the Human Genome Project (to study human DNA) was being planned, it was thought
that the task may require 100 years. However, technological change in the areas of computers, robotic sequencers, and the internet after the world-wide-web appeared in
1991 (to coordinate the activities of a large number of universities where each one was assigned a small piece of the problem), allowed the human genome project to
publish results after only 15 years (a 660% increase).
- Distributed computing projects like Folding@home and BOINC
have only been possible since 1995:
- the world-wide-web (proposed in 1989 to solve a document sharing problem among scientists at CERN in Geneva; then implemented in 1991) began to make the internet both popular and ubiquitous.
- CISC was replaced with RISC which further evolved to superscalar RISC then multicore.
- Vector processing became ubiquitous (primarily) in the form of video cards.
Processor technology was traditionally defined this way:
- Scalar (one data stream per instruction. e.g. CISC CPU)
- Superscalar (1-6 non-blocking scalar instructions simultaneously in a pipeline. e.g. RISC CPU)
- See: Flynn's Taxonomy for definitions like SISD (single instruction single data) and SIMD (single instruction multiple data) but remember that Data represents "Data stream"
- See: Duncan's taxonomy for a more modern twist
Caveat: these lists purposely omit things like SMP (symmetric multiprocessing) and VAX Clusters
Then CISC and RISC vendors began adding vector processing instructions to their CPU chips which blurred everything:
- Vector Processing (multiple data streams per instruction)
- Terminology from math, science and computer science:
- scalar: any measurement described by one data point (e.g. 30 km/hour)
- vector: any measurement described by two data points (e.g. 30 km/hour, North)
- A collection of vectors is usually referred to as a matrix (although a 2-dimensional data structure created in a computer is also known as a matrix; this includes a single spreadsheet as well as a set of database records where the columns represent field names; note that these examples can contain scalars, vectors, and tensors)
- tensor: any item involving three, or more, data points.
- Vector processing is a generic name for any kind of multi data point math (vector or tensor) performed on a computer.
- Tensor programming is not new in computing. Climate modelling begins with weather-prediction trials on ENIAC (a scalar machine) in 1947
- Google released a neat math library in 2015 called TensorFlow
- Technological speed up:
- While it is possible to do floating point (FP) math on integer-only CPUs, adding specialized logic to support FP and transcendental math can decrease FP processing time by one order of magnitude (x10) or more.
- Similarly, while it is possible to do vector processing (VP) on a scalar machine, adding specialized logic can decrease VP processing time by 2 to 3 orders of magnitude (x100 to x1000).
- Terminology from math, science and computer science:
Development over the decades:
- Mainframe Computers
- Minicomputer / Workstation
- 1989: DEC (Digital Equipment Corporation) adds vector processing to their Rigel uVAX chip
- 1989: DEC adds optional vector processing to VAX-6000 model 400 minicomputer
- http://manx-docs.org/collections/mds-199909/cd1/vax/60vaaom1.pdf (VAX 6000 Series - Vector Processor Owner’s Manual)
- https://manx-docs.org/collections/mds-199909/cd1/vax/60vaapg1.pdf (VAX 6000 Series - Vector Processor Programmer’s Manual)
- http://www.1000bit.it/ad/bro/digital/djt/dtj_v02-02_1990.pdf (Digital Technical Journal - Vol-2-Num-2 - Spring 1990)
- http://bitsavers.informatik.uni-stuttgart.de/pdf//datapro/datapro_reports_70s-90s/DEC/M11-325-50_9201_DEC_VAX6000.pdf (DataPro 1992)
- comments:
- The VAX-6000 was the Chevy of the computer industry during this time, and I worked on a VAX-6430 at my place of employment (here, 3 in the tens position meant 3 scalar CPUs). The cheapest new VAX-6000 varied in price between US$120k and US$750K depending upon model and options.
- While I never saw a vector processor board at my place of employment, I did see one at a DEC training facility in Maynard Massachusetts. They were only available for VAX-6410 and VAX-6510, and cost US$26K (which seems like a lot of money for a board that only offered two vector processors)
- 1994: VIS 1 (Visual Instruction Set) was introduced into UltraSPARC processors by Sun Microsystems
- comment: UltraSPARC was a 64-bit implementation of 32-bit SPARC.
- 1996: MDMX (MIPS Digital Media eXtension) is released by MIPS.
- 1997: MVI (Motion Video Extension) was implemented on the Alpha 21164. MVI appears again in Alpha 21264 and Alpha 21364.
- http://www.alphalinux.org/docs/MVI-full.html
- archive: https://web.archive.org/web/20140909020709/http://www.alphalinux.org/docs/MVI-full.html
- comment: Alpha was a 64-bit RISC successor to VAX.
- Microcomputer / Desktop
- 1997: MMX was implemented on P55C (a.k.a. Pentium 1) from Intel
and introduced 57 MMX-specific instructions.
- 1998: 3DNow! was implemented on AMD K-2.
- 1999: AltiVec (also called "VMX" by IBM and "Velocity Engine" by Apple) was implemented on PowerPC 4 from Motorola.
- 1999: SSE (Streaming SIMD Extensions) was implemented on Pentium 3 "Katmai" from Intel.
- 2001: SSE2 was implemented on Pentium 4 from Intel
- 2004: SSE3 was implemented on Pentium 4 Prescott on from Intel
- 2006: SSE4 was implemented on Intel Core and AMD K10
- 2008: AVX (Advanced Vector Instructions) proposed by Intel + AMD
but not seen until 2011.
- many components extended to 256-bits.
- 2012: AVX2 (more components extended to 256-bits)
- 2015: AVX-512 (512-bit extensions) first proposed in 2013 but not seen until 2015
- many components extended to 512-bits.
Technology Width Year MMX 64 bits 1997 SSE 128 bits 1999 AVX 256 bits 2008 AVX-512 512 bits 2015 - 1997: MMX was implemented on P55C (a.k.a. Pentium 1) from Intel
and introduced 57 MMX-specific instructions.
- Add-on graphics cards
- GPU (graphics programming unit) takes vector processing to a whole new level. Why? A $200.00 graphics card now can equip your system with 1500-2000 streaming processors and 2-4 GB of additional high-speed memory. According to the 2013 book "CUDA Programming", the author provides evidence why any modern high-powered PC equipped with one, or more (if your motherboard supports it), graphics cards can outperform any supercomputer listed 12 years ago on www.top500.org
- Many companies manufactured graphics cards (I recall seeing them available as purchase options in the IBM-PC back in 1981) but I will only mention two
companies here:
- ATI Technologies (founded in 1985)
- introduces GPU chipsets in the early 1990s that can do video processing without the need for a CPU.
- introduces the Radeon line in 2000 specifically targeted at DirectX 7.0 3D acceleration.
- acquired by AMD in 2006.
- ATI Technologies (founded in 1985)
- The circle of life?
- specialized mainframe computers from companies like IBM and Cray are built to host many thousands of "non-video video cards" (originally targeted for PCs and workstations). IBM's Roadrunner is one example.
To learn more:
- https://en.wikipedia.org/wiki/Graphics_processing_unit (GPU)
- https://en.wikipedia.org/wiki/GPGPU (General Purpose computing on Graphics Processing Units)
- Math + Science:
- https://en.wikipedia.org/wiki/CUDA
- https://en.wikipedia.org/wiki/OpenCL
- folding at home (protein analysis)
- http://chortle.ccsu.edu/VectorLessons/vectorIndex.html (Vector Math Tutorial)
- https://en.wikipedia.org/wiki/Tensor_processing_unit (a Google TPU is built using ASICs)
- Nvidia science cards (graphics cards without a video connector) break Moore's Law every year
- 2015 Maxwell_(microarchitecture)
- 2016 Pascal_(microarchitecture)
- 2017 Volta_(microarchitecture) and Turing_(microarchitecture)
- https://www.theregister.co.uk/2017/05/24/deeper_dive_into_gtc17/
- 2020 Ampere_(microarchitecture)
- 2022 Hopper_(microarchitecture) and Ada_Lovelace_(microarchitecture)
- 2024 Blackwell_(microarchitecture)
- 2028 coming: Feynman and Rubin
- typical core counts in 2025:
- eBooks
- Programming on Parallel Machines (V1.4 2014)
- see chapter 5 for CUDA examples in the C programming language.
- Is Parallel Programming Hard, And, If So, What Can You Do About It? (2017)
- https://developer.nvidia.com/gpugems/gpugems3/part-vi-gpu-computing
- Programming on Parallel Machines (V1.4 2014)
- Graphics:
- https://en.wikipedia.org/wiki/DirectX
- https://en.wikipedia.org/wiki/Direct3D (When the history of computing is written, Microsoft will be better known for DirectX and Direct3D than Windows)
- https://en.wikipedia.org/wiki/OpenGL
- Gaming
- Both the PlayStation 4 as well as the XBOX One employ an 8-core APU (Accelerated Processing Unit) made by AMD code-named Jaguar. What is an APU? It is a multi-core CPU with an embedded Graphics Chip Engine. Placing both systems on the same silicon die eliminates the signal delay associated with sending signals over an external bus.
- The PlayStation 5 employs an 8-core SoC made by AMD code-named Oberon. What is an SoC? It is a multi-core CPU integrated with an GPU.
- How do Video Game Graphics Work? (2024) https://www.youtube.com/watch?v=C8YtdC8mxTU
- Distributed computing projects like Folding@home and BOINC have only been
practical since 2005 when the CPUs in personal computers began to out-perform mini-computers and enterprise servers. Partly because...
- AMD added 64-bit support to their x86 processor technology calling it x86-64. (Linux distros still refer to this as a 686)
- Intel followed suit calling their 64-bit extension technology EM64T
- DDR2 memory became popular (this dynamic memory is capable of double-data-rate transfers)
- DDR3 memory became popular (this dynamic memory is capable of quadruple-data-rate transfers)
- Since then, the following list of technological improvements has made computers both faster and less expensive:
- Intel's abandonment of NetBurst which meant a return to shorter instruction pipelines starting with Core2
comment: AMD never went to longer pipelines; a long pipeline is only efficient when running a static CPU benchmark for marketing purposes - not running code in real-world where i/o events interrupt the primary foreground task (science in our case) - multi-core (each core is a fully functional CPU) chips from all manufacturers.
- continued development of optional graphic cards where CPUs would off-load much work to a graphics co-processor system (each card provided hundreds to thousand
streaming processors)
- ATI Radeon graphics cards (ATI was acquired by AMD in 2009)
- Nvidia GeForce graphics cards
- development of high performance "graphics" memory technology (e.g. GDDR3 , GDDR4 , GDDR5) to bypass processing stalls caused when streaming processors are too fast.
- Note that GDDR5 is used as main memory in the PlayStation 4 (PS4). While standalone PCs were built to host an optional graphics card, it seems that Sony has flipped things so that their graphics system is hosting an 8-core CPU. These hybrids go by the name APU (Accelerated Processing Unit)
- shifting analysis from host CPU cores (usually 2-4) to thousands of streaming processors
- Intel replacing 20-year old FSB technology with a proprietary new approach called QuickPath Interconnect (QPI) which is now found in Core-i3, Core-i5, Core i7 and Xeon
Historical note:- DEC created the 64-bit Alpha processor which was first announced in 1992 (21064 was first, 21164, 21264, 21364, came later)
- Compaq bought DEC in 1998.
- The DEC division of Compaq created CSI (Common System Interface) for use in their EV8 Alpha processor which was never released.
- HP merged with Compaq in 2002.
- HP preferred Itanium2 (jointly developed by HP and Intel) so announced their intention to gracefully shut down Alpha.
- Alpha technology (which included CSI) was immediately sold to Intel.
- Approximately 300 Alpha engineers were transferred to Intel between 2002 and 2004.
- CSI morphed into QPI (some industry watchers say that Intel ignored CSI until the announcement by AMD to go with a new industry-supported technology known as HyperTransport.
- The remainder of the industry went with a non-proprietary technology called HyperTransport which has been described as a multi-point Ethernet for use within a computer system.
- Intel's abandonment of NetBurst which meant a return to shorter instruction pipelines starting with Core2
- As is true in any "demand vs. supply" scenario, most consumers didn't need additional computing power which meant that chip manufacturers had to drop their prices just to keep the computing marketplace moving. This was good news for people setting up "folding farms". Something similar is happening today with computer systems since John-q-public is shifting from "towers and desktops" to "laptops and pads". This is causing the price of towers and graphics cards to plummet ever lower. You just can't beat the price-performance ratio of a Core-i7 motherboard hosting an NVIDIA graphics card.
- Shifting from brute-force "Chemical Equilibrium" algorithms to techniques involving Bayesian statistics and Markov Models will enable some exponential speedups.
- Computational Chemistry
Student Questions:- Using information from the periodic table of the elements you can see that the molecular mass of water (H2O) is ~18 which is lighter than many gases so why is water in a liquid state at room temperature while other slightly heavier molecules take the form of a gas?
- Ethanol (a liquid) has one more atom of Oxygen than Ethane (a gas). How can this small difference change the state?
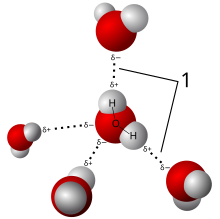 Liquid Water
Liquid Water
This diagram depicts an
H2O molecule loosely
bound to four others by
Van der Walls forces.Substance Molecule Atomic
MassesMolecular
MassState at Room
TemperatureWater H2O (1x2) + 16 18 liquid Carbon Monoxide CO 12 + 16 28 gas Molecular Oxygen O2 (16x2) 32 gas Carbon Dioxide CO2 12 + (16x2) 44 gas Ozone O3 (16x3) 48 gas Methane CH4 12 + (1x4) 16 gas Ethane C2H6 (12x2) + (1x6) 30 gas Ethanol C2H6O (12x2) + (1x6) + 16 46 liquid Propane C3H8 (12x3) + (1x8) 44 gas Butane C4H10 (12x4) + (1x10) 58 gas Pentane C5H12 (12x5) + (1x12) 72 gas Hexane C6H14 (12x6) + (1x14) 86 liquid Heptane C7H16 (12x7) + (1x16) 100 liquid Octane C8H18 (12x8) + (1x18) 114 liquid Short answers:- In the case of an H20 (water) molecule, even though two hydrogen atoms are covalently bound to one oxygen atom, those same hydrogen atoms are also attracted to each other which causes the water molecule to bend into a Y shape (according to VSEPR Theory). At the mid-point of the bend, a positive electrical charge from the oxygen atom is exposed to the world which allows a weak connection to the hydrogen atom of a neighboring H20 molecule (water molecules weakly sticking to each other form a liquid). These weak connections are called Van der Waals forces
- Below are the molecular schematic diagrams of Ethane (symmetrical) and Ethanol (asymmetrical). Notice that Oxygen-Hydrogen kink dangling to the right side of
Ethanol. That kink is not much different than a similar one associated with water. That is the location where a Van der Waal force weakly connects with an adjacent
ethanol molecule (not shown). So it should be no surprise that ethane at STP (Standard
Temperature and Pressure) exists as a gas while Ethanol exists as a liquid.
Ethane Ethanol (symmetrical) (asymmetrical) H H H H H | | | | / H-C-C-H H-C-C-O | | | | H H H H
Chemistry Caveat: The Molecular Table above was only meant to get you thinking. Now inspect this LARGER periodic table of the elements where the color of the atomic number indicates whether the natural state is solid or gaseous:
- all elements in column 1 (except hydrogen) are naturally solid.
- all elements in column 8 (helium to radon) are naturally gaseous.
- half the elements in row 2 starting with Lithium (atomic number 3) and ending with Carbon (atomic number 6), as well as two thirds of row 3 starting with Sodium (atomic number 11) and ending with Sulfur (atomic number 16), are naturally solid. I will leave it to you to determine why.
- Molecular Dynamics
Proteins come in many shapes and sizes. Here is a very short list:
These molecules are so large that modeling their interactions can only be done accurately with a computer.Protein Mass Function Notes Chlorophyll a 893 facilitates photosynthesis in plants Heme A 852 common ligand for many hemeproteins including hemoglobin and myoglobin Alpha-amylase 56,031 salivary enzyme to digest starch pdbId=1SMD hemoglobin 64,458 red blood cell protein DNA polymerase varies from
50k to 200kenzyme responsible of DNA replication

FAH Links
| https://foldingathome.org | cover page |
| https://foldingathome.org/home/ | home page |
| https://foldingathome.org/start-folding/ | download page |
| https://foldingforum.org | official technology problems, discussions, news, science, etc. |
| https://www.reddit.com/r/Folding/ | unofficial technology problems, discussions |
| https://en.wikipedia.org/wiki/Folding@home | Wikipedia article |
FAH Targeted Diseases
This "folding knowledge" will be used to develop new drugs for treating diseases such as:
- ALS ("Amyotrophic Lateral Sclerosis" a.k.a. "Lou Gehrig's Disease")
- Alzheimer's Disease
- Plaques, which contain misfolded peptides called amyloid beta, are formed in the brain many years before the signs of this disease are observed. Together, these plaques and neurofibrillary tangles form the pathological hallmarks of the disease.
- Cancer & P53
- P53 is the suicide gene involved in apoptosis (programmed cell death - something necessary in order your immune system to kill cancer cells)
- CJD (Creutzfeldt-Jakob Disease)
- the human variation of mad cow disease
- Huntington's Disease
- Huntington's disease is caused by a trinucleotide repeat expansion in the Huntingtin (Htt) gene and is one of several polyglutamine (or PolyQ) diseases. This expansion produces an altered form of the Htt protein, mutant Huntingtin (mHtt), which results in neuronal cell death in select areas of the brain. Huntington's disease is a terminal illness.
- Osteogenesis Imperfecta
- Normal bone growth is a yin-yang balance between osteoclasts and oseteoblasts. Osteogenesis Imperfecta occurs when bone grows without sufficient or healthy collagen (protein)
- Parkinson's Disease
- The mechanism by which the brain cells in Parkinson's are lost may consist of an abnormal accumulation of the protein alpha-synuclein bound to ubiquitin in the damaged cells.
- Ribosome & antibiotics
- A ribosome is a protein producing organelle found inside each cell.
Reference Links: Folding@home - FAQ Diseases
More Information About Proteins and Protein-Folding Science
- Pande Science - Home
- What is "The Pande Group"?
- History of Folding@Home, Vijay Pande
- Why study, simulate, use Folding@Home, Vijay?
- What are the basic protein structures?
- Protein functions in the body
- Electrostatics and protein folding
- When and where do proteins fold?
- How does entropy affect protein folding?
- What are chaperones and how do they work?
Online Documents
- Wikipedia
- AlphaFold by DeepMind (a Google Alphabet Company)
- https://en.wikipedia.org/wiki/AlphaFold
- https://www.deepmind.com/research/highlighted-research/alphafold/timeline-of-a-breakthrough
- While not part of the folding@home project, AlphaFold involves protein folding.
- Their project took 5 years to design then implement.
- They mapped all 200 million known proteins then freely published it as a gift to humanity.
- Assuming that one human researcher (a PhD) could do one protein in 5-years, then AlphaFold saved 1 billion-man-years of time.
- https://alphafold.ebi.ac.uk/ <<<--- access their online protein database
- Science (2020-11-30) https://www.science.org/content/article/game-has-changed-ai-triumphs-solving-protein-structures
- Nature (2020-11-30) https://www.nature.com/articles/d41586-020-03348-4
- Nobel Prize in Chemistry (2024-10-09) https://www.nobelprize.org/prizes/chemistry/2024/press-release/ (Protein Folding)
- Nobel Prize in Physics (2024-10-08) https://www.nobelprize.org/prizes/physics/2024/press-release/ (Neural Network path to modern A.I.)
- AlphaFold 2 (2021-07-19)
- AlphaFold 3 (2024-05-08)
- Cornell University
- https://arxiv.org/abs/2303.08993 Folding@home: achievements from over twenty years of citizen science herald the exascale era. (a.k.a. 10^18 FLOPS)
- How AI Cracked the Protein Folding Code and Won a Nobel Prize (22-min video)
- Veritasium: The Most Useful Thing AI Has Done (Protein folding: from Rosetta@home to DeepMind)
https://www.youtube.com/watch?v=P_fHJIYENdI
With an overview of CASP (Critical Assessment of Structure Prediction) from CASP1 (1994) to CASP13 (2018) and beyond - We Solved the Protein Folding Problem… Now What? (StarTalk + Neil deGrasse Tyson)
https://www.youtube.com/watch?v=hJ4LRswmZEs
My Computational Statistics (with some help from China)
- BOINC stats:
- https://www.boincstats.com/stats/-1/user/detail/206514146972/projectList
Name: Neil Steven Rieck
Projects: Rosetta@Home, Docking@Home, POEM@Home, SETI@home
- https://www.boincstats.com/stats/-1/user/detail/206514146972/projectList
- FAH official stats:
- https://stats.foldingathome.org/team/3213 (team: China Folding@Home Power) Top 100 Ranked Team
- https://stats.foldingathome.org/team/10987 (team: Canada) Top 1K Ranked Team
- https://stats.foldingathome.org/donor/75502 (user: neil_rieck) Top
1K Ranked Donor world rank: 397 (as of 2026-02-27)
- 12,929,000,000 points
- Too many zeroes? This number is currently over 12 billion.
- I started in 2007 with a half-dozen PCs (running Windows) and one PlayStation 3; I am now running a half-dozen PCs (running Linux Mint) hosting Nvidia graphics cards as compute engines.
- 160,000 work units
- 12,929,000,000 points

- FAH third-party stats:
- https://folding.lar.systems
- Many like-minded people in China are helping with protein folding (this is great news). Some of their client processes are folding under my account name at a very respectable 10 million points per day. Whoever you are, many thanks for helping humanity advance biological knowledge. Isaac Asimov would approve.
from to user
team third party statics rank 2007-12-22 2022-01-28 neil_rieck Default Final Account Tally ( Points: 418,381,428 WU: 110,984 )
https://folding.extremeoverclocking.com/user_summary.php?s=&u=306663
Note: no new stats since 2022-01-28 because I changed teams1005
2022-01-28 present neil_rieck Canada https://folding.extremeoverclocking.com/user_summary.php?s=&u=1291088
11
Canada https://folding.extremeoverclocking.com/team_summary.php?s=&t=10987 94 2023-03-04 present neil_rieck China Folding@Home Power https://folding.extremeoverclocking.com/user_summary.php?s=&u=1316605 12
China Folding@Home Power https://folding.extremeoverclocking.com/team_summary.php?s=&t=3213 12
Folding via GPUs (graphics cards)
- A single core Pentium-class CPU provides one scalar processor. It also provides one streaming/vector processor under marketing names like MMX (64-bit) , SSE (128-bit), AVX (256-bit) and AVX-512 (512-bit)
- Most Core i5 and Core i7 CPU's provide four scalar processors so also offer at least four streaming/vector processors.
- However, a single add-on graphics card can also provide hundreds to thousands of streaming/vector processors (the Nvidia RTX-3090 provides 10,496)
Folding with Linux (2019 - 2025)
also click: Linux tips: real world problems and solutionsFolding with EL7 (2019)
I just found two junk PCs in my basement with 64-bit CPUs that were running 32-bit operating systems (Windows-XP and Windows-Vista). Unfortunately for me, neither were eligible for Microsoft's free upgrade to Windows-10, and I had no intention of buying a new 64-bit OS just for this hobby, so I swapped out Windows with CentOS-7 and was able to get each one folding with very little effort. What follows are some tips for people who are not Linux gurus:
- Download a DVD ISO image of CentOS-7 via this link: https://www.centos.org/download/
- CentOS-7.7 and CentOS-8.0 were released days apart in September 2019 (perhaps due to the invisible hand of IBM?)
- Linux distros assume you will copy these images to a USB stick, but the BIOS in many older machines does not support booting from a thumb drive.
- Software from the top of download page preferentially offers CentOS-8 which is too large (> 4.7 GB) to write to a single-layer writable DVD so chose CentOS-7
- Note: I have had some success with Dual Layer optical media but all your drives needs to support it.
- Transfer to bootable media (choose one of the following)
- copy the ISO image to a DVD-writer
-OR- - use rufus to format an USB stick then copy the ISO image to the USB
caveat: newer PCs have transitioned from BIOS to UEFI. Older BIOS-based systems do not support booting from a USB stick (strange because you can connect a USB-based DVD then boot from that)
- copy the ISO image to a DVD-writer
- Boot-install CentOS-7 on the 64-bit CPU
- Using a larger downloaded image:
1) burn, boot, install Linux using recipe: "GUI with Server"
2) reboot; now update via the internet like so: yum update
3) reboot; 4) add development tools: yum group install "Development Tools" - Using smaller downloads (then adding GUI after the fact):
1) burn, boot, install Linux using recipe: "Server"
2) reboot; now update via the internet like so: yum update
3) reboot; optionally enable GUI: yum group install "Server with GUI"
reboot; then type...
systemctl isolate graphical.target
systemctl set-default graphical.target 4) add development tools: yum group install "Development Tools"
- Using a larger downloaded image:
- My machines hosted Nvidia graphics cards (GTX-560 and GTX-960 respectively)
so these systems required the correct Nvidia drivers in order to do GPU-based folding. Why? The generic drivers only support video but folding science requires OpenCL and CUDA
- HARD WAY: If you are a Linux guru and know how to first disable the Nouveau driver, then install the 64-bit Linux driver provided by NVIDIA here: http://www.nvidia.com/Download/index.aspx
- EASY WAY: First install the nvidia-detect module found at elrepo (Enterprise Linux REPOsitory) here ( http://elrepo.org/tiki/tiki-index.php
) and documented here ( https://elrepo.org/tiki/nvidia-detect ) then use the utility to install another elrepo
package. The steps look similar to this if you are logged in as root:
step-1 (install one/two repos via yum)
activity Linux command Notes Description Notes view available repos yum list \*-release\*
backslash escapes the asterisk
install elrepo yum install elrepo-release required Enterprise Linux REPO (hobbyist supported) nvidia-detect is found here install epel yum install epel-release optional Extra Packages for Enterprise Linux (RedHat supported) httpd and mysql are found here
step-2 (install nvidia-detect from elrepo)
If you are not logged in as root then you must prefix every command with sudo (super user do).activity Linux command Notes install nvidia-detect yum install nvidia-detect
test nvidia-detect nvidia-detect -v
install nvidia driver yum install $(nvidia-detect) BASH passes the output of nvidia-detect to yum reboot reboot
Now jump to: Linux v7 common
Folding with EL8 (2022 - 2024)
caveats:
- EL is a popular internet acronym for Enterprise Linux which includes RHEL, CentOS, AlmaLinux, Rocky Linux, Oracle Linux, EuroLinux (and sometimes SEL, SUSE and OpenSUSE). These distros employ packages with a file extension of ".rpm" (redhat package manager) so are referred to as rpm-based.
- Debian, Ubuntu, and Linux Mint (to only name a few of many) employ packages with a file extension of "deb" (Debian) so are referred to as deb-based.
- While it is still possible to do GPU folding on EL8 platforms like RHEL-8 in 2025 via the old v7 client, the folding@home project is moving to a v8 client which (as
of 2025-07-20 with v8.4) only supports deb-based distros but not rpm-based distros. I suspect the folding@home developers made this decision because:
- IBM purchased Red Hat then ended point releases of CentOS
- obtaining prebuilt Nvidia drivers with "CUDA baked in" is virtually impossible with EL8 (perhaps EL distros were never intended to be used as desktops)
update: On 2025-08-06 the AlmaLinux maintainers announced on Reddit that Alma will be providing package support for "Nvidia hardware drivers with CUDA" for both AlmaLinux-9 and AlmaLinux-10. So people more comfortable with EL platforms might wish to give AlmaLinux a try (I have it running on more than a dozen large HPE servers at my employer's data center) - building your own Nvidia driver with CUDA is too difficult for the majority of people wishing to contribute to folding@home
- First off, new FAH downloadable cores require an updated version of Linux library glibc which is not available on CentOS-7, so you need to upgrade to CentOS-8, or any other EL8 (see note #5 below)
- Secondly, changes to the FAH GPU cores now require a minimum of OpenCL-1.2. This means that my GTX-560 is no longer useful as a streaming processor. I noticed another blurb about double-precision FP math which definitely rules out my GTX-560 so I replaced it with a GTX-1650
- On both systems I replaced CentOS-7 with CentOS-8 (via a fresh install) then followed the CentOS-7 instructions just above. However, the Nvidia driver from
elrepo-release (EL8) no longer contains any support for OpenCL or CUDA, so I was forced to install the Linux driver provided by Nvidia
- you will first need to disable the Nouveau video driver which requires some Linux guru voodoo (see below)
- For some reason, these CentOS-8 machines seemed sluggish at the time, so I jumped to Rocky Linux 8.5 which fixed my problem. Since then I have come to believe that AlmaLinux would be a better choice.
- Caveat: IBM purchased Red Hat in 2019 for the sum of US$34 billion. In 2020 the resultant Blue Hat
announced their intent to discontinue CentOS-8 minor point updates after 2021-12-31 because too many companies (like Facebook and Twitter) were using the free version
rather than paying for RHEL (gotta speed up the ROI on that $34 billion investment). Even though Red Hat begins each OS iteration using open-source software, on 2023-06-21 they announced that they were going to restrict access (close source) their modifications. This will affect
all EL (Enterprise Linux) flavors of Linux including: AlmaLinux, EuroLinux, Oracle Linux, Rocky Linux, etc. so you might consider moving to any target supported by
this migration tool (ELevate - leapp). Note that academics working at Fermilab just outside of Chicago, or CERN
(home of the LHC) in Geneva, recommend moving to AlmaLinux and I agree. My advice is to always
employ a Linux that can be downloaded from a university mirror. Here is one example of 10,000: https://mirror.csclub.uwaterloo.ca/
Mirror observations (2024):Real world concern: you never know when problems (political, commercial, technological) will block you from getting to a single site. But you can always modify "/etc/yum.conf" to point directly to a nearby university mirror (if files exist on the university mirror; most mirrors now only contain AlmaLinux offerings)
- active offerings exist only for AlmaLinux (this appears to be the only EL available on the university mirror)
- all CentOS folders (point release and stream) are empty
- no mirror offerings for: EuroLinux, Oracle Linux, or Rocky Linux
- Updating to a NVIDIA published driver
- after the initial Linux install, type "sudo yum update" to bring the platform up to the latest level. If a new kernel was installed then you must reboot before you continue.
- now type "sudo yum install elrepo-release"
- now type "sudo yum install nvidia-detect"
- now type "nvidia-detect -v" so you know what Nvidia driver is required for the current Nvidia graphics card.
- DO NOT use elrepo to update the Nvidia driver (on 2022-01-16 it was missing support for both OpenCL
and CUDA)
update 2025-07-31:- while the Nvidia drivers for Windows automatically install support for CUDA, this does not appear to be the case for CentOS-8 or anything higher.
- you might wish to use elrepo to update the Nvidia driver provided you can also locate and install this package nvidia-cuda-toolkit
- alternatively, you might wish to try installing cuda-toolkit directly from Nvidia: https://developer.nvidia.com/cuda-downloads?target_os=Linux
- alternatively, visit here: https://rpmfusion.org/Howto/CUDA
- If all this seems too much of a pain, then you might wish to try a friction-free install via Linux Mint
- read these reference notes to download/build an Nvidia driver with CUDA support:
- http://www.nvidia.com/object/unix.html (choose the file indicated by "nvidia-detect -v")
- https://linuxconfig.org/install-the-latest-nvidia-linux-driver
- https://linuxconfig.org/how-to-install-the-nvidia-drivers-on-centos-8
- https://support.huawei.com/enterprise/en/doc/EDOC1100165479/93fe5683/how-to-disable-the-nouveau-driver-for-different-linux-systems (many thanks to my computer buddies in China for this last tip)
- steps:
- Download the desired driver file from NVIDIA into the root account
- for GTX960 I used: NVIDIA-Linux-x86_64-470.86.run (will probably be changed before you read this)
- for GTX1650 I used: NVIDIA-Linux-x86_64-525.89.run (will probably be changed before you read this)
- Now disable the Nouveau driver
- create file /etc/modprobe.d/blacklist-nouveau.conf containing these two lines:
blacklist nouveau
options nouveau modeset=0 - create a new ramdisk for use during system boot: dracut --force
- reboot
- caveat: at this point your monitor is no longer capable of displaying small text in character-cell mode but you don't care because you've already downloaded the necessary files from Nvidia corporation. Right?
- create file /etc/modprobe.d/blacklist-nouveau.conf containing these two lines:
- Install the NVIDIA driver
- yum group install "Development Tools"
- chmod 777 NVIDIA-Linux-x86_64-470.86.run
- ./NVIDIA-Linux-x86_64-470.86.run
- caveat: the previous command might fail for the following reasons:
- not an executable file (did you use chmod ?)
- no build tools (gcc compiler, etc. Did you install "Development Tools"?)
- nouveau driver is still running
- caveat: the previous command might fail for the following reasons:
- reboot
- Now click: Common to all v7 client installs on Linux
- Download the desired driver file from NVIDIA into the root account
- caveats:
- kernel updates via "yum update" (CentOS-7) or "dnf update" (CentOS-8) always require a reboot. Some updates to the kernel may clobber the Nvidia driver (so just repeat step 3 above).
- If your console is blank (oops!) then type this three-key-combo CTL-ALT-F3 (control alternate F3) then log in as root on the third virtual console. You will be working from the command line.
Folding with Ubuntu (2025)
caveats:
(1) While the various Enterprise Linux (EL) version numbers tend to move in lockstep, this is not true with non-EL Linux versions like Ubuntu (or distros derived from
it like Linux Mint). At the time of this writing, EL-8-9-10 are all currently being covered by active support. Meanwhile, a new version of Ubuntu is released every two
years, and the version number is based upon the current year. At the time of this writing, than version number is 24.04 (released April of 2024)
(2) While it appears that the Linux desktop user community is currently in love with Ubuntu, I have run into several annoying problems with this OS which prevent FAH
clients (both v7 + v8) from running autonomously across the 6 PCs in my basement. This may have something to do with snap (something that is disabled by default in Linux Mint) which is constantly trying to update software even after I attempted to disable this
feature. On top of that, snap is definitely slowing down my GUI-based app startup. So click: Linux Mint 2025
- Download the Desktop version of Ubuntu from here: https://ubuntu.com/download/desktop
- Since many of my older machines do not support booting from a thumb drive, I needed to copy the ISO to optical media
- I download an optical media burner from here: https://www.nch.com.au/burn/index.html (will
require a double-sided DVD in order to write a 6-GB ISO file)
- click here for more help: Linux Notes: ISO images
- connected a USB-based ASUS (SDRW-08D2S-U) Slim External 8x DVD Writer
- CAVEAT: this drive is power hungry so both USB connectors (on the drive) must be plugged in for both reading as well as writing.
- One of my systems did not have two adjacent USB ports so I had to buy a so-called USB power bar. Twenty bucks on Amazon.
- I download an optical media burner from here: https://www.nch.com.au/burn/index.html (will
require a double-sided DVD in order to write a 6-GB ISO file)
- Boot the optical media (or USB stick (thumb drive))
- choose "Try or Install Ubuntu" (Ubuntu will be loaded into memory)
- be patient. Wait until you see both of these icons on your desktop:
- "Home"
- "Install Ubuntu 24.04.02 LTX"
- Double click on the icon "Install Ubuntu 24.04.02 LTX"
- answer 12 questions
- on question 8 you should enable checkbox "install third-party software for graphics and Wi-Fi hardware" in order to get the proper Nvidia video driver which includes CUDA
- answer 12 questions
- bring your OS up to date via the Terminal command line
$ sudo apt update # update your local cache
$ sudo apt upgrade #
$ sudo reboot #
Using the v8.4 fah client
- Download a Linux FAH client: https://download.foldingathome.org/ (it will be dropped into your Downloads sub-folder)
- Install your Linux FAH client
- click on the "show apps" icon
- click on "Terminal"
$ pwd
/home/neil
$ cd Downloads
$ pwd
/home/neil/Downloads
$ sudo apt install ./fah-client_8.4.9_amd64.deb
{{{ verbage ensues }}}
{{{ tips are displayed }}}
$ systemctl --no-pager -l fah-client
$ ----------------------------------- if not running then continue
$ sudo systemctl fah-client start
- Configure your local Linux FAH client (via your browser: https://v8-4.foldingathome.org/).
- Click on the gear icon for account settings like Username + Team
- This is also the place where you can "set the CPU count to zero" and "enable your GPU"
- The icon beside the gear will allow you to view the machine log for the client in question
- Optionally, read the manual: https://foldingathome.org/guides/v8-4-client-guide/
- Observations: I replaced Windows-10 with Ubuntu-24 so am running the exact same hardware.
- It would appear that the FAHClient is two-to-three times faster under Ubuntu-24 than Windows-10
(how could this be? is Windows that much slower -OR- did Windows reserve too many streaming processors for its own use?)
- It would appear that the FAHClient is two-to-three times faster under Ubuntu-24 than Windows-10
- STOP HERE (you are done)
Using the older v7 client
When the v8 client runs into trouble (e.g. can't get a work unit, etc) it waits for user input before proceeding which can be a pain if you are running 6 machines like me. So you might wish to go back and install a v7 client. I tried this on 2025-07-29 and it works so click: Common to all v7-client installs on Linux to see how to do it.
Folding with Linux Mint (2025 - 2026)
- Download the Cinnamon version of Linux Mint from here: https://linuxmint.com/download.php (at the time of this writing it is version 22.02)
- Since many of my older machines do not support booting from a thumb drive, I needed to copy the ISO to optical media
- I download an optical media burner from here: https://www.nch.com.au/burn/index.html (will
require a double-sided DVD in order to write a 2-GB ISO)
- click here for more help: Linux Notes: ISO images
- connected a USB-based ASUS (SDRW-08D2S-U) Slim External 8x DVD Writer
- CAVEAT: this drive is power hungry so both USB connectors (on the drive) must be plugged in for both reading as well as writing.
- One of my systems did not have two adjacent USB ports so I had to buy a so-called USB power bar. Twenty bucks on Amazon.
- I download an optical media burner from here: https://www.nch.com.au/burn/index.html (will
require a double-sided DVD in order to write a 2-GB ISO)
- Boot the optical media (or USB stick (thumb drive))
- choose "Start Linux Mint" (Linux Mint will be loaded into memory)
- be patient. Wait until you see this icon on your desktop: install Linux Mint
- double click on: Install Linux Mint
- bring your OS up to date via the Terminal command line
$ sudo apt update # update your local cache
$ sudo apt upgrade #
$ sudo reboot # - disable the update manager which is run at startup
- System Settings >> Startup Applications: disable Update manager
- change your video driver from nouveau to Nvidia
- Driver Manager: take note of the hardware installed (mine: GeForce RTX 3050 6GB)
- Driver Manager: change from "xserver-xorg-video-nouveau" to "nvidia-driver-575" (you might wish to double check this last number by doing a driver search at Nividia)
- click: Restart
- install CUDA support via the Terminal command line
$ sudo apt update # update your local cache (again)
$ sudo apt install nvidia-cuda-toolkit #
$ sudo reboot #
- Now click: Common to all v7 client installs on Linux (v7 is good enough for Linux Mint; v8 is currently a waste of time)
Common to all v7 client installs on Linux
- Linux software an be downloaded from here:
- https://download.foldingathome.org/ (page of supported software; v8 is at the top, v7 is at the bottom)
- https://download.foldingathome.org/releases/ (okay to browse)
- Installing Folding-at-home on Linux
- some of the GUI-based software may not work properly (on CentOS-7 or earlier) because Python3 is a prerequisite
- Python3 can be easily added to EL7 systems like CentOS-7 but do not remove or disable Python2 because that action will break certain system utilities like yum or firewall-cmd to name two of many.
sudo yum install epel-release
sudo yum install python36 - Python-3.6.8 is the standard offering in EL8 offerings like RHEL-8, AlmaLinux-8, etc.
- Python3 can be easily added to EL7 systems like CentOS-7 but do not remove or disable Python2 because that action will break certain system utilities like yum or firewall-cmd to name two of many.
- So I recommend:
- only install the command-line client as described here: https://foldingathome.org/support/faq/installation-guides/linux/manual-installation-advanced/
- perform a manual configure as described here: https://foldingathome.org/support/faq/installation-guides/linux/command-line-options/
- starting the client with the --configure switch will generate an XML configuration file
- starting the client with the --config switch will let you test an XML configuration file
- starting the client with the --help switch will display more help than you ever dreamed possible
- Caveat: just installing the FAH-Client will cause it to be installed as a service then start CPU folding (which is
probably what you do not want). If you want to enable GPU-based folding (and/or disable CPU folding) then you will need to stop the client, modify the
config file, test the config file, then restart the client. Here are some commands to help out.
task command stop service sudo systemctl status | grep -i fah # Enterprise Linux (EL)
sudo systemctl stop FAHClient # Enterprise Linux (EL)
or
sudo ps -efH | grep -i fah # UNIX or Linux
sudo /etc/init.d/FAHClient stop # UNIX or Linuxnavigate here cd /etc/fahclient edit config file
(nano does not
require vi or
vim skills)vi config.xml
or
nano config.xml
<config>
<power v='full'/>
<user v='neil_rieck'/>
<team v='10987'/>
<gpu v='true'/>
<smp v='false'/>
<cpu v='false'/>
<slot id='1' type='GPU'/>
</config>
notes:
1) client type can be any of: 'cpu', 'smp', 'gpu'
2) never enable 'smp' unless to have >= 4 cores
3) with 'gpu' enabled I see no point in folding with 'cpu' or 'smp'navigate here cd /var/lib/fahclient (optional) /usr/bin/FAHClient --help (optional) /usr/bin/FAHClient --info
note: Nvidia GPU's are not visible here because they are managed by CUDAinteractive testing /usr/bin/FAHClient --config /etc/fahclient/config.xml -v
note: GPU folding requires OpenCL-1.2 so if you see errors like
'cannot find OpenCL' then you might need to rebuild your NVIDIA
driver (almost always required after any Linux update that
replaces the kernel)end interactive test hit: <ctrl-c> start the service systemctl start fahclient
or
sudo /etc/init.d/FAHClient start &monitor step #1
monitor step #2
monitor step #3top -d 0.5
cat /var/lib/fahclient/logs.txt
tail -40 /var/lib/fahclient/logs.txt
Solving a v7 no-WU problem sudo systemctl stop FAHClient # stop the v7 client
cd /var/lib/fahclient # move to this v7 folder
sudo rm -fr * # delete all files here and below
sudo systemctl start FAHClient # start the v7 client
tail logs.txt # ensure work units are being downloadedSolving a v8 no-WU problem sudo systemctl stop fah-client # stop the v8 client
cd /var/lib/fah-client # move to this v8 folder
sudo rm -fr * # delete all files here and below
sudo systemctl start fah-cleint # start the v8 client
tail logs.txt # ensure work units are being downloaded
- some of the GUI-based software may not work properly (on CentOS-7 or earlier) because Python3 is a prerequisite
Microsoft Windows Scripting and Programming
- MS-DOS/MSDOS Batch Files: Batch File Tutorial and Reference
- MS-DOS @wikipedia
- Batch file @wikipedia
- Microsoft Windows XP - Batch files
Experimental Stuff for Windows Hackers
DOS commands for creating, and starting, a Windows Service to execute a DOS script.
sc create neil369 binpath="cmd /k start c:\folding-0\neil987.bat" type=own type=interactsc start neil369
Once created, you can stop/start/modify a service graphically from this Windows location:
Start >> Programs >> Administrative Tools >> ServicesStopped services may only be deleted from DOS like so:
sc query neil369sc delete neil369
BOINC (Berkeley Open Infrastructure for Network Computing)
- If you are unable to pick a single cause then pick several because the BOINC manager will switch between science clients every hour (this interval is adjustable). In my case I actively support POEM, Rosetta, and Docking.
- The current BOINC client can be programmed to use one, some, or all cores of a multi-core machine. The BOINC client can also utilize (or not) the streaming processors on your Graphics Card.
Protein / Biology / Medicine Projects
General
POEM@home (via BOINC)
- https://en.wikipedia.org/wiki/POEM@Home
- https://www.youtube.com/watch?v=EchPDQRRYq4 (promotional German-language video with English subtitles)
Rosetta@home (via BOINC)
- http://boinc.bakerlab.org/rosetta/ is the home of Rosetta@home
which operates through the BOINC framework. Their graphics screen-saver is one very effective way to help visualize "what molecular dynamics is all about". All
science teachers must show this to their students.
- I'm sure everyone already knows that a computer "rendering beautiful graphical displays" is doing less science. That said, humans are visual creatures and graphical displays have their place in our society. Except for some public locations, all clients should be running in non-graphical mode so that more system resources are diverted to protein analysis.
- Five questions for Rosetta@home: How Rosetta@home helps cure cancer, AIDS, Alzheimer's, and more
- https://en.wikipedia.org/wiki/Rosetta@home
- https://www.youtube.com/watch?v=GzATbET3g54 (official 7 minute video at YouTube.com)
World Community Grid (via BOINC)
- http://www.worldcommunitygrid.org - sponsored by IBM
- http://www.wcgrid.org/join - use this link to access the WCG-specific BOINC client.
Notes:- some people may prefer to use the generic BOINC client from Berkley then install the WCG plugin from that application; you will still need to create your WCG account at the WCG site
- You only need to do this if you want to cycle your BOINC client between multiple projects of which WCG is just one
- If you only want to run the WCG project (which also switches between IBM sponsored science projects) then it probably makes more sense to use the WCG-specific client
- https://en.wikipedia.org/wiki/World_community_grid (WCG) is an effort to create the world's largest public computing grid to tackle scientific research projects that benefit humanity. Launched 2004-11-16, it is funded and operated by IBM with client software currently available for Windows, Linux, Mac-OS-X and FreeBSD operating systems. They encourage their employees and customers to do the same.
- http://www.worldcommunitygrid.org/research/proteome/overview.do - Human Proteome Folding
- http://www.worldcommunitygrid.org/research/hpf2/overview.do - Human Proteome Folding - Phase 2
Personal Comment: I wonder why HP (Hewlett-Packard) has not followed IBM's lead. Up until now I always thought of IBM as the template of uber-capitalism but it seems that the title of "king of profit by the elimination of seemingly superfluous expenses" goes to HP. Don't they realize that IBM's effort in this area is done under IBM's advertising budgets? Just like IBM's 1990s foray into chess playing systems (e.g. Deep Blue) led to increased sales as well as share prices, one day IBM will be able to say "IBM contributed to a treatments for human diseases including cancer". IBM actions in this area reinforce the public's association with IBM and information processing.
Biology Science Links
- Howard Hughes Medical Institute
- Research Collaboratory for Structural Bioinformatics (RCSB)
- http://www.rcsb.org/pdb/101/structural_view_of_biology.do - Structural View of Biology
- http://www.biomachina.org/research/
- Custom-Built Supercomputer Brings Protein Folding Into View
http://biomachina.org/publications_web/SHAW10_news_of_the_week.pdf
- Custom-Built Supercomputer Brings Protein Folding Into View
- Introduction to Protein Folding and Molecular Simulation - Tokyo University of Science - Tadashi Ando
Protein Data Bank Links
- http://www.pdb.org/pdb/101/motm.do?momID=31 p53 Tumor Suppressor
- http://www.pdb.org/pdb/101/motm.do?momID=78 Luciferase
- http://www.pdb.org/pdb/101/motm.do?momID=74 Alpha-amylase (Salivary Digestive Enzyme)
- http://www.pdb.org/pdb/101/motm.do?momID=41 Hemoglobin
- http://www.pdb.org/pdb/101/motm.do?momID=11 Rubisco
- http://www.pdb.org/pdb/101/motm.do?momID=3 DNA Polymerase
ENCODE
The Encyclopedia of DNA Elements (ENCODE) Consortium is an international collaboration of research groups funded by the National Human Genome Research Institute (NHGRI). The goal of ENCODE is to build a comprehensive parts list of functional elements in the human genome, including elements that act at the protein and RNA levels, and regulatory elements that control cells and circumstances in which a gene is active.- http://genome.ucsc.edu/ENCODE/
- https://en.wikipedia.org/wiki/ENCODE
- The Encyclopedia of DNA Elements (ENCODE) is a public research consortium launched by the US National Human Genome Research Institute (NHGRI) in September 2003. The goal is to find all functional elements in the human genome, one of the most critical projects by NHGRI after it completed the successful Human Genome Project. All data generated in the course of the project will be released rapidly into public databases.
- On 5 September 2012, initial results of the project were released in a coordinated set of 30 papers published in the journals Nature (6 publications), Genome Biology (18 papers) and Genome Research (6 papers). These publications combine to show that approximately 20% of noncoding DNA in the human genome is functional while an additional 60% is transcribed with no known function. Much of this functional non-coding DNA is involved in the regulation of the expression of coding genes. Furthermore, the expression of each coding gene is controlled by multiple regulatory sites located both near and distant from the gene. These results demonstrate that gene regulation is far more complex than previously believed.
- http://www.nature.com/encode/
- http://www.nature.com/nature/journal/v489/n7414/full/489052a.html - ENCODE Explained
- http://www.nature.com/nature/journal/v489/n7414/full/489049a.html - The Making of ENCODE
- http://www.nature.com/news/encode-the-human-encyclopaedia-1.11312 - What’s next for ENCODE?
- Bits of Mystery DNA, Far From ‘Junk,’ Play Crucial Role (what was once called "junk DNA" is now referred to as "dark genetic material"
http://www.nytimes.com/2012/09/06/science/far-from-junk-dna-dark-matter-proves-crucial-to-health.html - http://silvertonconsulting.com/blog/2012/09/07/big-sciencebig-data-encode-project-decodes-junk-dna
Local Links
- Genes as Technology 1 - comparing genes to computers (information storage)
- Genes as Technology 2 - comparing genes to computers (error detection and correction)
- Guaranteed Human Life Extension - not a joke or scam, but it will cost you $6.00 per month.
(noteworthy) Remote Links
- https://www.youtube.com/watch?v=lNh0Km4bv18&NR=1 - An introduction to protein structure and function.
-
https://www.khanacademy.org/science/biology/evolution-and-natural-selection/v/dna - An introduction to DNA
- https://www.youtube.com/watch?v=w8VOfmG985U - DNA Lesson - Khan Academy
- https://www.khanacademy.org/science/biology/cell-division/v/chromosomes--chromatids--chromatin--etc - Chromosomes, Chromatids, Chromatin, etc.
- 2013
- http://www.technologyreview.com/view/510571/the-million-core-problem/ The
Million-Core Problem - Stanford researchers break a supercomputing barrier.
quote: A team of Stanford researchers have broken a record in supercomputing, using a million cores to model a complex fluid dynamics problem. The computer is a newly installed Sequioa IBM Bluegene/Q system at the Lawrence Livermore National Laboratories. Sequoia has 1,572,864 processors, reports Andrew Myers of Stanford Engineering, and 1.6 petabytes of memory. - http://www.wired.com/wiredenterprise/2013/01/million-core-supercomputer/ Researchers Set Record With Million-Core Calculation
- http://www.technologyreview.com/view/510571/the-million-core-problem/ The
Million-Core Problem - Stanford researchers break a supercomputing barrier.
- 2020
- 2024
- 2025
- Veritasium: The Most Useful Thing AI Has Done (Protein folding from Rosetta@home to DeepMind)
https://www.youtube.com/watch?v=P_fHJIYENdI
With an overview of CASP (Critical Assessment of Structure Prediction) from CASP1 (1994) to CASP13 (2018) and beyond
- Veritasium: The Most Useful Thing AI Has Done (Protein folding from Rosetta@home to DeepMind)
Recommended Biology Books (I own them all)
- The Eighth Day of Creation (1979/1993/1996/2004) Horace
Freeland Judson - highly recommended (25th anniversary edition)
- starts with DNA; ends with RNA to Amino Acid mapping in ribosomes.
- The Code of Codes (1992/2000) Daniel Kevles and Leroy Hood
- subtitled: Scientific and Social Issues in the Human Genome Project
- Epigenetics (2011) Richard C Francis
- how our environment enables/disables/modulates DNA expression.
- The Epigenetics Revolution (2012) Nessa Carey
- how our environment enables/disables/modulates DNA expression.
- Life at the Speed of Light (2013) J. Craig Venter
- synthetic biology: from computers to DNA.
 Back to Home
Back to HomeNeil Rieck
Waterloo, Ontario, Canada.
